Nov 30, 2017
DeepNeuro

A 3D Convolutional Neural network iteratively learns to annotate a tumor. (gif by me, from a model developed in DeepNeuro). Github link here.
DeepNeuro comes with pre-trained networks for performing segmentation of glioblastoma (particularly aggressive brain tumors), melanoma metastases in the brain (often smaller, more numerous and homogenous brain tumors), stroke lesions, and segmentation of brain tissue (an essential preprocessing step for downstream analyses). All of these networks are made available via Docker containers, and can be run via command-line utilities in Python. Tutorials have been developed in Google Colab to facilitate this process.
For those who wish to train their own algorithms, DeepNeuro has pre-built functions that allow inexperienced or time-crunched users to get started with a minimum of training. These include pre-built functions for image segmentation (U-Net), image classification, and image sythesis (PGGAN). Data-loaders for the confusing variety of medical imaging data formats are available, and pre-processing and data augmentation functions specific to radiology are also included. Functions for evaluating the accuracy of users' algorithms and visualizing training progress are also available.
I developed DeepNeuro while working as a research asssitant and then programmer at the Massachusetts General Hospital (MGH). I worked in the Quantitative Translational Imaging Lab (QTIM) supervised by Dr. Jayashree Kalpathy-Cramer, Dr. Elizabeth Gerstner, and Dr. Bruce Rosen. Myself and members of our lab have used DeepNeuro to do research on several areas of radiology, including research on glioblastoma, melanoma metastases, adrenoleukodystrophy, ischemic stroke, radiation therapy, and other projects. At the time of my departure from MGH, we were testing DeepNeuro with collaborators in MGH and out, including the Tata Memorial Hospital in India and the MD Anderson Cancer Center in Houston.
DeepNeuro is still under development at QTIM with the lab's current staff. The next challenge for DeepNeuro, other than building additional pre-trained networks and machine learning functions, is to bridge the gap between deep learning and actual clinical practice. This involves many steps to make the package easier to use for lay-users and technical support teams, such as graphical user interfaces and server-mode. Collaborators the Center for Clinical Data Science (CCDS) at Massachusetts General Hospital, and well as technicians in MGH itself can help us bridge that gap.
You can see an arxiv paper for DeepNeuro here. An updated paper is currently undergoing revisions in a peer-reviewed journal, which hopefully I'll get to soon :). The following papers have used DeepNeuro, or involve code developed for DeepNeuro:
- Beers, Andrew, Ken Chang, James Brown, Elizabeth Gerstner, Bruce Rosen, and Jayashree Kalpathy-Cramer. 2018. “Sequential Neural Networks for Biologically Informed Glioma Segmentation.” In Medical Imaging 2018: Image Processing, 10574:1057433. International Society for Optics and Photonics. link.
- Chang, Ken, Andrew L. Beers, Harrison X. Bai, James M. Brown, K. Ina Ly, Xuejun Li, Joeky T. Senders, et al. 2019. “Automatic Assessment of Glioma Burden: A Deep Learning Algorithm for Fully Automated Volumetric and Bi-Dimensional Measurement.” Neuro-Oncology, June. https://doi.org/10.1093/neuonc/noz106.
- Beers, A., J. Brown, K. Chang, and J. P. Campbell. 2018. “High-Resolution Medical Image Synthesis Using Progressively Grown Generative Adversarial Networks.” arXiv Preprint arXiv. https://arxiv.org/abs/1805.03144.
- Chang, K., J. Brown, A. Beers, J. Kalpathy-Cramer, and H. Ay. 2019. “Viscerotoxic Brain Infarcts: The Results of Heart-Brain Interactions Study.” Stroke; a Journal of Cerebral Circulation. https://www.ahajournals.org/doi/abs/10.1161/str.50.suppl_1.162.
- Silva, Michael A., Jay Patel, Vasileios Kavouridis, Troy Gallerani, Andrew Beers, Ken Chang, Katharina V. Hoebel, et al. 2019. “Machine Learning Models Can Detect Aneurysm Rupture and Identify Clinical Features Associated with Rupture.” World Neurosurgery 131 (November): e46–51. link.
- Bakas, S., M. Reyes, A. Jakab, and S. Bauer. 2018. “Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge.” arXiv Preprint arXiv. https://arxiv.org/abs/1811.02629.
- Winzeck, Stefan, Arsany Hakim, Richard McKinley, José A. A. D. S. R. Pinto, Victor Alves, Carlos Silva, Maxim Pisov, et al. 2018. “ISLES 2016 and 2017-Benchmarking Ischemic Stroke Lesion Outcome Prediction Based on Multispectral MRI.” Frontiers in Neurology 9 (September): 679. link.
Some older screenshots of DeepNeuro's outputs can be found below.
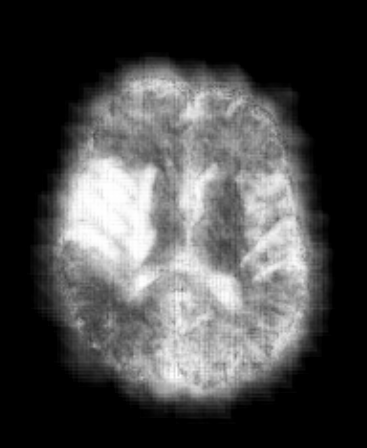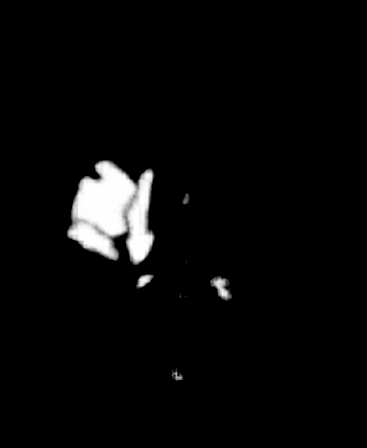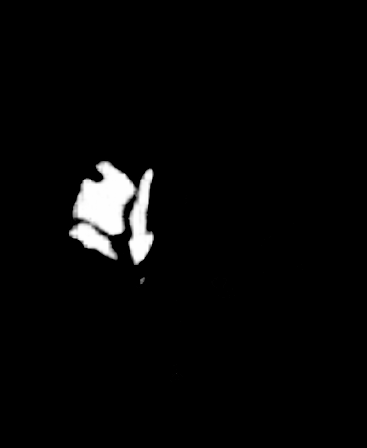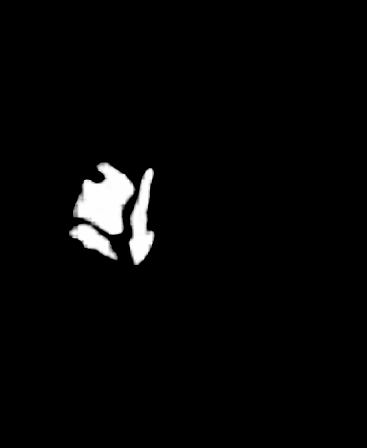
A 3D Convolutional Neural network iteratively learns to annotate a tumor. (gif by me, from a model developed in DeepNeuro).
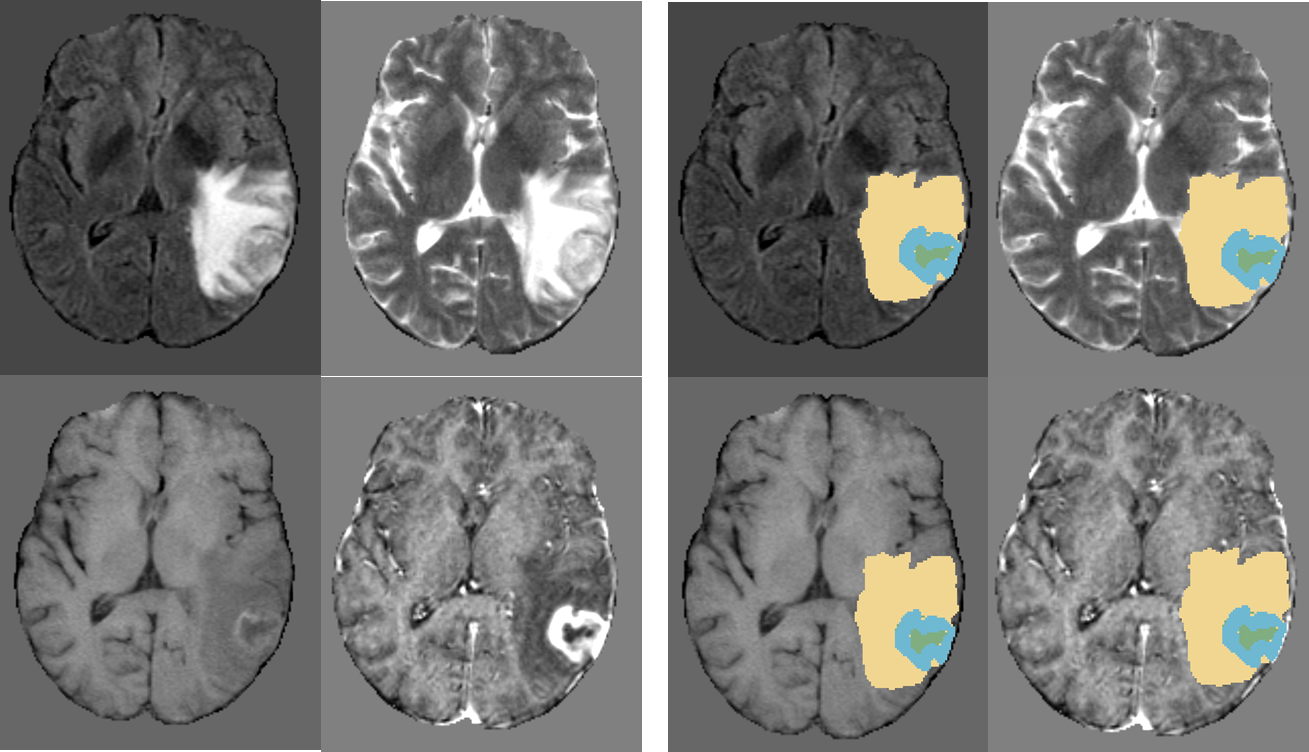
Results of DeepNeuro on a sample glioblastoma case. Enhancing tumor is labeled blue, peritumoral edema is labeled yellow, and necrotic tissue is labeled green.